The original article was written by Jen Sheen, and this page was made by Hirokazu Kobayashi but not copied from an online journal site to avoid copyright infringement.
The following Greek characters used may be correctly displayed with Western
fonts such as "Helvetica", "Times", etc.: "µ" for "micro", and "‡" for "omega".
Engineered GFP as a vital reporter in plants
Background: The green fluorescent protein (GFP) of the jellyfish Aequorea
victoria has recently been used as a universal reporter in a broad range of
heterologous living cells and organisms. Although successful in some plant
transient expression assays based on strong promoters or high copy number viral
vectors, further improvement of expression efficiency and fluorescent intensity are
required for GFP to be useful as a marker in intact plants. Here, we report that an
extensively modified GFP is a versatile and sensitive reporter in a variety of living
plant cells and in transgenic plants.
Results: We show that a re-engineered GFP gene sequence, with the favored
codons of highly expressed human proteins, gives 20-fold higher GFP expression
in maize leaf cells than the original jellyfish GFP sequence. When combined with a
mutation in the chromophore, the replacement of the serine at position 65 with a
threonine, the new GFP sequence gives more than 100-fold brighter fluorescent
signals upon excitation with 490 nm (blue) light, and swifter chromophore
formation. We also show that this modified GFP has a broad use in various
transient expression systems, and allows the easy detection of weak promoter
activity, visualization of protein targeting into the nucleus and various plastids, and
analysis of signal transduction pathways in living single cells and in transgenic
plants.
Conclusions: The modified GFP is a simple and economical new tool for the
direct visualization of promoter activities with a broad range of strength and cell
specificity. It can be used to measure dynamic responses of signal transduction
pathways, transfection efficiency, and subcellular localization of chimeric proteins,
and should be suitable for many other applications in genetically modified living
cells and tissues of higher plants. The data also suggest that the codon usage
effect might be universal, allowing the design of recombinant proteins with high
expression efficiency in evolutionarily distant species such as humans and maize.
Background
The green fluorescent protein (GFP) of the jellyfish Aequorea victoria has a number
of desirable traits as a universal reporter in living cells and organisms [1,2]. Apart
from an apparent requirement for molecular oxygen, the formation of the
fluorophore appears to be cell-autonomous [1-3]. Direct visualization of gene
expression in individual cells is therefore possible without cell lysis and
subsequent biochemical analysis, and tissue distortion caused by fixation, staining
and section can be avoided. The autocatalytic formation of the chromophore and
relative resistance to photobleaching make GFP an attractive fluorescent tag for
studying protein interaction, localization and traffic [2-5]. Although the expression of
GFP has been demonstrated in Escherichia coli, yeast, Caenorhabditis elegans,
Drosophila, mammals and plants [1-15], the broader application of GFP in
mammals and higher plants requires higher expression efficiency and fluorescent
intensity, especially under blue light, to minimize photobleaching and phytotoxicity
[2,6]. Here, we have used a modified GFP, and show that it acts as a convenient
and sensitive reporter for the visualization of gene regulation, signal transduction
and subcellular localization of chimeric proteins in living cells of maize, tobacco,
onion and Arabidopsis, and in transgenic tobacco plants.
Results
Expression of engineered GFP in plant cells
As each GFP molecule represents one fluorophore, high-level expression is
important to give good fluorescent signals. Using universal transcription and
translation enhancers with strong promoters, we have shown previously that GFP
signals are detectable in transient expression systems of monocot maize and dicot
Arabidopsis [12,13]. However, an increase in the expression efficiency and
quantum yield with 490 nm excitation would make GFP substantially more useful as
a vital marker in plants. As the preferred codon usage is almost identical between
humans and maize, and is compatible with that of other higher plants (such as
Arabidopsis) [16,17], we tested the expression of a new synthetic GFP gene
sequence (sGFP) with optimal human codons [17] in plant cells. A mutation in the
chromophore the replacement of the serine position 65 with a threonine (S65T) has
been shown in E. coli to result in enhanced brightness, faster chromophore
formation and slower photobleaching [7]. This change was also introduced into
sGFP by site-directed mutagenesis, to create sGFP(S65T).
Three similar constructs were generated by inserting GFP, sGFP or sGFP(S65T)
into a plant expression vector with a strong, constitutive promoter (35SC4PPDK)
and the 3' NOS transcription terminator [12,18]. Plasmid DNA was introduced into
maize leaf protoplast by electroporation. After 3-4 hours, bright fluorescence
signals were visible under blue light in 50 % of protoplast transfected with the
35SC4PPDK-sGFP(S65T) construct (Fig. 1a-c). This early detection of sGFP(S65T)
was due mainly to the enhanced fluorescent signal and rapid chromophore
formation induced by the S65T mutation, as shown in E. coli [7]. After 15-16 hours
of incubation, green fluorescence was detectable in 50 % of all transfected
protoplast, although the fluorescent intensity was much stronger in cells
electroporated with the sGFP and sGFP(S65T) constructs (Fig. 1d-f). As GFP and
sGFP have identical amino-acid sequences, the brighter signal from sGFP is likely
to be due to a higher level of protein synthesis caused by optimal codon usage, as
demonstrated by Haas et al. in the preceding paper [17]. To confirm that the
selected codon usage gave higher protein expression in maize leaf cells, we
examined the amount of 35S-methionine-labeled GFP expressed by the three
constructs. As shown in Figure 1g, the amounts of newly synthesized sGFP and
sGFP(S65T) were similar and easily detectable without purification, whereas the
amount of native GFP was about 20-fold lower.
----------------------------------------------------
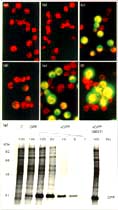
Figure 1
Engineered GFP gives faster and brighter fluorescent signals. Maize mesophyll
protoplast were electroporated with three plasmids expressing (a,d) the jellyfish
GFP (GFP), (b,e) synthetic GFP (sGFP) or (c,f) a mutant S65T sGFP (sGFP(S65T)).
The transfected protoplast were observed after (a-c) 4 h or (d-f) 1 6 h of incubation.
(g) Synthetic GFP gives a high level of protein expression. Untransfected control
(C) and transfected maize protoplast were labeled with 400 µCi ml-1
[35S]methionine for 12 h. Total proteins were solubilized in protein loading buffer
and separated by 12.5% SDS-PAGE.
----------------------------------------------------
sGFP(S65T) as a reporter for a weak promoter
As the fluorescent intensity of sGFP(S65T) was dramatically improved, we tested its
use as a reporter for a heterologous weak promoter. Activity of the dicot
Arabidopsis CAB2 promoter (AtCAB2) [19,20] could be detected when fused to
luciferase (LUC) [21-24] and chloramphenicol acetyltransferase (CAT) [25,Z6] (data
not shown), but not when native GFP was used in maize leaf cells [12]. An AtCAB2
sGFP(S65T) construct was electroporated into monocot maize leaf cells, and the
cells incubated with or without light; after 20 hours incubation, bright fluorescence
was only detected in the cells incubated under light (Fig. 2). The sensitivity of
sGFP(S65T) as a reporter appeared comparable to that of CAT and LUC. In spite of
its lower activity, the Arabidopsis CAB2 promoter is regulated by light in a similar
manner to the maize photosynthetic gene promoters [18,26-28]. This indicates the
presence of a universal light signal transduction pathway in dicot and monocot leaf
cells.
----------------------------------------------------

Figure 2
The expression of AtCAB2-sGFP(S65T) is regulated by light in maize leaf
protoplast. (a) Untransfected control protoplast. (b,c) Transfected protoplast
incubated (b) in the dark or (c) under light for 20 h.
----------------------------------------------------
sGFP(S65T) as a vital reporter in tobacco protoplast
We have shown previously that GFP expression can be detected in a maize
protoplast transient expression system. In tobacco protoplast, however, it was
difficult to visualize GFP expression from the native GFP sequence even with a
strong promoter. Maize leaf protoplast appear to have substantially more synthetic
capacity than protoplast isolated from leaves of a number of plant species [25 28].
To use GFP in leaf protoplast from tobacco and many other plant species therefore
requires a higher level of GFP expression. A construct carrying 35SC4PPDK
sGFP(S65T) was introduced into tobacco leaf protoplast by polyethylene glycol
(PEG)-mediated transfection [29]. As shown in Figure 3, bright green fluorescence
was detected in > 80% of the protoplast after 20 hours of incubation (Fig. 3).
Weaker signals were also obtained with a similar construct carrying 35SC4PPDK
sGFP (data not shown). Tobacco leaf protoplast are very sensitive to hormone
treatment and can easily undergo dedifferentiation and regeneration [30]. In
combination with the sGFP(S65T) marker, these cells can be used conveniently to
study signal transduction and cell-cycle regulation in higher plants [30].
----------------------------------------------------
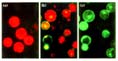
Figure 3
The expression of 35SC4PPDK-sGFP(S65T) in tobacco mesophyll protoplast. (a)
Untransfected control and (b,c) transfected tobacco protoplast after 20 h incubation.
In (c) the red autofluorescence of chlorophyll was blocked using a interference
filter.
----------------------------------------------------
Analysis of organelle targeting in Arabidopsis
Many plant proteins have to be targeted to the nucleus and various plastids to
serve their proper functions. To visualize such targeting, we used sGFP(S65T)
chimeric proteins with either the nuclear localization sequence (NLS) [31] or the
plastid transit peptide (TP) [32]. Plasmid constructs were introduced into both living
leaves and roots of Arabidopsis by DNA bombardment, and the targeting
sequences directed the localization of sGFP(S65T) into the nucleus and plastid
(Fig. 4b,c,e,f). Without a targeting sequence, sGFP(S65T) accumulated diffusely in
the cytoplasm and nucleus (Fig. 4a,d). As no manipulation was required prior to
sample observation, the integrity of cell structure and morphology was maintained
perfectly. Although plastids in roots and leaves have very different morphologies
and functions, the signals and machineries for protein import seem to bc similar.
The sGFP(S65T) reporter appears to be superior to ß-glucuronidase (GUS) fusions
[33], which are larger, demand exogenous substrate and infiltration, need cell and
tissue fixation, and have leakage problems because the indigo dye generated by
enzymatic action often precipitates diffusely. It provides a new and powerful visual
tool to study organelle targeting in living cells and to select mutants with abnormal
protein localization in intact plants.
----------------------------------------------------
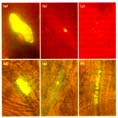
Figure 4
Organelle targeting in Arabidopsis. Constructs carrying (a,d) 35S‡-sGFP(S65T),
(b,e) 35S‡-NLS-sGFP(S65T) or (c,f) 35S‡-TP-sGFP(S65T) were bombarded into
Arabidopsis (a-c) leaves or (d-f) roots. The expression and localization of
sGFP(S65T) was observed after 24 h of incubation.
----------------------------------------------------
Nuclear targeting in onion epidermal cells
Onion skin epidermal cells have recently become a popular system in the study of
regulation and sequence requirements for nuclear localization in plants [34].
Constructs expressing sGFP(S65T) and NLS-sGFP(S65T) were bombarded into
epidermal cells. Striking signals were observed after 24 hours of incubation,
demonstrating the use of this new marker in a simple system (Fig. 5).
----------------------------------------------------
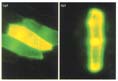
Figure 5
Nuclear localization of sGFP(S65T) in onion cells. Constructs carrying (a) 35S‡
sGFP(S65T) or (b) 35S‡-NLS-sGFP(S65T) were bombarded into onion skin
epidermal cells. The expression and localization of sGFP(S65T) was observed
after 24 hours incubation.
----------------------------------------------------
A favorable reporter for identification of transgenic plants
Inefficient expression and low brightness of the native GFP have impeded its use
as a visual reporter in transgenic plants. Using sGFP(S65T) and sGFP (data not
shown) fused to the 35SC4PPDK promoter, transgenic tobacco plants were easily
identified by examining directly the leaves of putative transformants under a
fluorescence microscope. Untransformed tobacco leaves appeared bright red (Fig.
6a), whereas transformed tobacco leaves showed yellow fluorescence, the
consequence of sGFP(S65T) expression (Fig. 6b). The red autofluorescence of
chlorophyll was easily blocked by an interference filter (Fig. 6c). This visual
identification of plants based on transgene expression is faster, simpler and more
reliable than drug selection and polymerase chain reaction (PCR)-based
identification. All protoplasts isolated from the leaves of transgenic plants showed
green fluorescence, so GFP expression was not limited to a particular cell type (Fig.
6d-f). Most sGFP(S65T) was concentrated around the plasma membrane, and
around or in the nucleus. The expression of sGFP(S65T) directed by cell-type
specific promoters will provide a simple and powerful means for sorting and
purifying various living cell types from transgenic plants using fluorescent-activated
cell sorting [12,13]. It should be noted that a very high level of GFP expression can
inhibit the segmentation process in producing transgenic plants (data not shown).
----------------------------------------------------
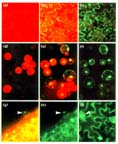
Figure 6
sGFP(S65T) as a vital marker in transgenic tobacco plants. (a,d) Untransformed
control and (b,c,e,f) transgenic tobacco plant (a-c) leaves and (d-f) protoplasts were
observed using a fluorescence microscope with a FITC filter set without or (c,f) with
an interference filter. (g-i) Drought-inducible expression of sGFP(S65T) in
transgenic tobacco plants. Transgenic plants carrying the AtRD29A-sGFP(S65T)
construct were allowed to wilt and the expression of sGFP(S65T) on leaf surface
was observed (g) without or (h,i) with an interference filter. Bright green
fluorescence was most striking in the nucleus of trichome and guard cells as
indicated by arrows.
----------------------------------------------------
Drought-inducible gene expression in transgenic plants
To illustrate the use of sGFP(S65T) as a reporter to study signal transduction in
intact plants, we generated transgenic tobacco plants carrying sGFP(S65T)
controlled by a drought-inducible promoter (RD29A) from Arabidopsis [35]. The
plants were allowed to wilt, to induce the expression of sGFP(S65T). Green
fluorescence was clearly visible in many cell types, including the epidermal,
mesophyll and guard cells of the leaves (Fig. 6g-i). Striking nuclear accumulation of
sGFP(S65T) was observed in guard cells and trichomes. No green fluorescence
was detected without the drought treatment. Thus, sGFP(S65T) can serve as an
excellent reporter allowing the direct visualization in real time of plant responses to
various environmental signals.
Discussion
The jellyfish GFP is a new reporter with high potential for use in all living cells and
organisms. Its use in mammals and higher plants, however, has been limited by
inefficient expression, low fluorescence, slow chromophore formation and complex
photoisomerization [2]. Several laboratories have made progress recently in
improving GFP to be a more versatile and sensitive reporter. These modifications
increase the sensitivity of GFP detection, reduce photo-bleaching and phototoxicity,
allow simultaneous analysis of two promoters or two proteins, and offer better
matching to standard fluorescence microscopy filter sets [2.3,7,8]. Higher levels of
GFP expression are still important to broaden its applications in mammals and
higher plants. By combining optimal codon usage [17] and the S65T mutation [7],
we have found that sGFP(S65T) gives higher expression levels, faster
chromophore formation and more enhanced fluorescent emission by blue light in
plant cells. Previously, a universal transcription or translation enhancer was
essential to use the original jellyfish GFP in monocot or dicot cells [12,13]. The
detection of GFP in tobacco leaves infected with the tobacco mosaic virus TMV
GFP [36] or the potato virus X PVY-GFP [9] relied on high copy numbers of TMV
and PVX RNA carrying the GFP sequence.
We have shown here that sGFP and sGFP(S65T) can be used to detect the activity
of weaker promoters. Furthermore, both work beautifully (without lethal staining,
fixation and dissection) in a broad spectrum of transient expression systems, such
as electroporated maize protoplasts, PEG-transfected tobacco protoplasts and
bombarded Arabidopsis and onion tissues. The expression of sGFP and
sGFP(S65T) also provides a rapid, simple and non-destructive assessment of
transient transfection and stable transformation effrciency. The incorporation of an
imaging system should add more precise quantitation power [24].
We have explored the use of sGFP(S65T) as a reporter for light- and drought
inducible gene expression mediated by universal signaling pathways in higher
plants. Both responses were conveniently visualized in individual cells. The
modified GFP will also be an invaluable reporter for monitoring plant responses to
other environmental stimuli, such as pathogens, wounding, touch and stresses, as
well as internal physiological, metabolic and developmental activities in living cells
and plants. This reporter can serve as a powerful tool, therefore, for elucidating the
mechanisms of gene regulation and signal transduction in higher plants.
The relatively low molecular weight of GFP makes it ideal as a fluorescent tag in
fusion proteins. Protein sorting, traffic, localization, intracellular fate and
extracellular movement can be followed in real time at high resolution, especially
with confocal microscopes [4,5,12,14]. Fusion proteins with various fluorescent tags
also provide a new way to detect protein-protein interactions or changes in protein
conformation in vivo. The sGFP(S65T) reporter is a convenient tool for further
studies at the single-cell level, and can label cells for functional and physiological
analysis of co-transfected genes. It also has potential as a vital marker for enhancer
and gene trap screening, investigating recombination and transposition events, fate
mapping or cell lineage analysis, tracing chromosome or gene segregation,
genetic and molecular mapping, and mutant selection.
Recently, Haseloff and colleagues [14] observed the splicing of a cryptic intron in
GFP mRNA which abolished GFP expression in transgenic Arabidopsis plants. It is
not clear how widespread this phenomenon is in other plant species. Our
observation of GFP expression in maize and Arabidopsis transient assays suggests
that GFP is not, or only partially, spliced in these cells. Splicing is unlikely to occur
in maize mesophyll protoplasts as a single polypeptide of the correct size [1,3,9,12]
is detected in cells transfected with GFP, sGFP and sGFP(S65T). Serendipitously,
the sequence at the cryptic splice donor site AAAGGTATTGATTTTAAA was
changed to AAgGGcATcGATTTcAAg during the synthesis of sGFP with favored
codons, and the cryptic intron in the coding region (400-483) of GFP [14] is
therefore eliminated in sGFP and sGFP(S65T).
Conclusions
We have shown that sGFP and sGFP(S65T) are versatile and sensitive reporters in
transient expression using maize, tobacco, onion and Arabidopsis cells, and in
transgenic tobacco plants. These new tools can be used for studies of gene
regulation, signal transduction, development and cell biology in higher plants.
Materials and methods
Plasmid Constructions
The construction of the plant expression vector with a strong and constitutive
promoter 35SC4PPDK has been described previously [1 2,18]. The creation of
sGFP is reported separately [1 7]. sGFP(S65T) was generated by PCR-based site
directed mutagenesis using two flanking primers: 5'
GCGGATCCATGGTGAGCAAG-3' and 5'-GGGCGGCCGCTTTACTTGTA-3' and two
overlapping mutagenesis primers: 5'-GTGACCACCTTCACCTACGGCGTGCAG-3'
and 5'-CTGCACGCCGTAGGGAAGGTGGTCAC-3'. GFP, sGFP and sGFP(S65T)
were amplified using the same flanking PCR primers and inserted into the
expression vector between BamHI and Smal sites. Three clones were picked from
each construction for initial evaluation The Arabidopsis CAB2 [19, 20] and RD29A
[35] promoters were obtained by PCR and fused to the Ncol site at the 5' of
sGFP(S65T). Three clones were selected for initial evaluation by transient
expression analysis. The primers used were 5'-TGGACTAGAGATTGCCACGTA-3'
and 5'-GGAGGAGAGAGCCATGGTTGAGGCGGCCAT-3' for the AtCAB2 promoter
and 5'-GACCGACTACTAATAATAGTAAGT-3' and 5'
TGTTTGATCCATGGTCCACCGATTTTT-3' for the AtRD29A promoter. The
constructs used for bombardment carried the 35S‡ regulatory sequence instead of
the 35SC4PPDK promoter [1 2]. The NLS of SV40 [31] was synthesized (5'
TCGACCATGGCTCCAAAGAAGAAGAGAAAGGT-3' and 5'
CATGACCTTTCTCTTCTTCTTTGGAGCCATGG-3'), annealed and kinased before
insertion into the SalI and Ncol site of the 35‡-sGFP(s65T) plasmid. The TP
sequence was obtained from RBCS-1A (-38 Bfal and Sphl +165, blunt ends) [32]
and inserted into the blunted SalI and NcoI site of the 35‡-sGFP(S65T) plasmid.
The binary vector used for tobacco transformation was pART27 [37], carrying
35SC4PPDK-sGFP(S65T) and AtRD29A-sGFP(S65T).
Protoplast transient expression
Maize seedlings were grown in the dark for 11-12 days before illumination for 16
18 h as described [25,26]. The preparation, electroporation and incubation of the
maize mesophyll protoplasts were as described [25,26]. Transfected protoplasts
were incubated at 23oC for 4-20 h to allow the accumulation of the GFP, sGFP and
sGFP(S65T). The labeling and analysis of GFP with 35S-methionine have been
described [1 2]. The protocols for tobacco mesophyll protoplasts preparation, PEG
transfection, and incubation were similar to those used for carrot protoplasts
described previously [29], with some modifications. Healthy and expanded
tobacco SR1 leaves were cut to about 2 cm2 and digested in an enzyme solution
consisting 1.2% Cellulase R10 and 0.4% Macerozyme R10 in K3 medium [38] with
0.4 M sucrose for overnight in the dark at 23oC. Protoplasts were collected by
floating. Plasmid DNA carrying 35SC4PPDK-sGFP(S65T) (20µg) was added to
0.25 ml freshly isolated tobacco protoplasts (106 ml-1) in 0.4 M mannitol, 20 mM
CaCl2, 5 mM MES, pH 5.7. An equal volume of 40% PEG 4000 in 0.4 M mannitol
and 100 µM Ca(NO3)2 (brought to pH 10 using KOH before autoclaving) was added
immediately, mixed well and incubated for 10 min at room temperature. The mix
was diluted with 4 ml K3 medium containing 0.3 M sucrose. The transfected
protoplasts were incubated in the dark for 20-24 h before being photographed.
Tissue Bombardment
Tissues from Arabidopsis thaliana (Columbia) were prepared as described
previously [12]. Onion epidermal cell layers were peeled and placed inside up on
the MS plates [12]. Plasmid DNAs of appropriate fusion genes (0.5 µg) were
introduced to Arabidopsis leaves and roots using a pneumatic particle gun (PDS
1000/He; BIO-RAD) The condition of bombardment was vacuum of 28 inch Hg,
helium pressure of 1550 or 1800 psi for Arabidopsis and 1100 or 1300 psi for
onion, and 6 cm of target distance using 1.1 µm of tungsten microcarriers. After
bombardment, tissues were incubated on the MS plates for 24 h at 22oC, Samples
were observed directly or transferred to glass slides.
Tobacco transformation
Stable transformation was performed based on the established protocol using
tobacco SR1 Leaves [39].
Fluorescence microscopy
The fluorescence photographs of maize mesophyll protoplasts were taken using a
Zeiss Universal microscope equipped with epifluorescence condenser III RS and a
FITC filter set comprising exciter filter (BP 450-490), chromatic beam splitter (FT
510), and barrier filter (LP 520), and Kodak Ektachome Elite 400 color film. The
optimal exposure time was 30 sec. The light source was provided by a HBO 50 W
high-pressure mercury bulb. The fluorescence photographs of tobacco protoplasts
and tissues were taken using a Leitz DM-R microscope through epifluorescence
filter set 13, which contains an excitation filter with band pass of 450-490 nm, RKP
510 dichromatic mirror, and 520 nm long pass filter. The microscope is also
equipped with an interference filter that can be used to block the red
autofluorescence from chlorophyll. The light source was provided by a 100 W high
pressure mercury bulb. Arabidopsis tissues were observed with Olympus
fluorescent microscopy (AH2-RFL) with a filter set providing 455-490 nm excitation
and emission above 515 nm.
References
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ: Primary
structure of the Aequora victoria green-fluorescent protein. Gene 1992, 111: 229
233.
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY: Understanding,
improving and using green fluorescent proteins. Trends Biochem Sci 1995 20:
448-455.
- Heim R, Prasher DC, Tsien RY: Wavelength mutations and posttranslational
autoxidation of green fluorescent protein. Proc Natl Acad Sci USA 1994, 91:
12501-12504.
- Wang S, Hazelrigg T: Implications for bcd mRNA Localization from spatial
distribution of exu protein in Drosophila oogenesis. Nature 1994, 369: 400-403.
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T: Chimeric green fluorescent
protein as a tool for visualizing subcellular organelles in living cells. Curr Biol 1995,
5: 635-642.
- Chalfie M Tu Y Euskirchen G, Ward WW, Prasher DC: Green fluorescent protein
as a marker for gene expression. Science 1994, 263: 802-805.
- Heim R, Cubitt AB, Tsien RY: Improved green fluorescence. Nature 1995, 373:
663-664.
- Delagrave S, Hawlin RE, Silva CM, Yang MM, Youvan DC: Red-shifted
excitation mutants of the green fluorescent protein. Biotechnology 1995, 13: 151
154.
- Baulcombe DC, Chapman S, Santa Cruz S: Jellyfish green fluorescent protein
as a reporter for virus infections. Plant J 1995, 7: 1045-1053.
- Niedz RP Sussman MR, Satterlee JS: GFP expression in transiently
transformed protoplasts made from nonphotosynthetic suspension culture of Citrus
sinensis. Plant Cell Report 1995, 14: 403-406.
- Hu W, Cheng CL: Expression of Aequorea green fluorescent protein in plant
cells. FEBS Lett 1995, 369: 331-334.
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW: Green-fluorescent
protein as a new vital marker in plant cells. Plant J 1995, 8: 777-784.
- Galbraith DW, Lambert GM, Grebenok RJ, Sheen J: Flow cytometry analysis of
transgene expression in higher plants: green-fluorescent protein. Meth Cell Biol
1995, 50: 3-14.
- Haseloff J, Amos B: GFP in plants. Trends Genet 1995, 11: 328-329.
- Yeh E, Gustafson K, Boulianne GL: Green fluorescent protein as a vital marker
and reporter of gene expression in Drosophila. Proc Natl Acad Sci USA 1995, 92:
7036-7040.
- Wada K, Wada Y, Doi H, Ishibashi F, Gojobori T, Ikemura T: Codon usage
tabulated from the GenBank genetic sequence data. Nuc Acid Res 1991, 19: 1981
1986.
- Haas J, Park E-C, Seed B: Codon usage limitation in the expression of HIV-1
envelope glycoprotein. Curr Biol 1996, 6: 315-324.
- Sheen J: Protein phosphatase activity is required for light-inducible gene
expression in maize. EMBO J 1993, 12: 3497-3505.
- Mitra A, Choi HK, An G: Structural and functional analyses of Arabidopsis
thaliana chlorophyll a/b-binding protein (cab) promoters. Plant Moi Biol 1989, 12:
169-179.
- Anderson SL, Kay S: Functional dissection of circadian clock-and
phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad
Sci USA 1995, 92: 1500-1504.
- Ow DW, Wood KV, DeLuca M, de Wet JR Helinski DR, Howell SH: Transient
and stable expression of the firefly luciferase gene in plant cells and transgenic
plants. Science 1986, 234: 856-859.
- Gallie DR, Lucas WJ, Walbot V:Visualizing mRNA expression in plant
protoplasts: factors influencing efficient mRNA uptake and translation. Plant Cell
1991, 1: 301-311.
- Luehrsen KR, de Wet JR, Walbot V: Transient expression analysis in plants
using firefly luciferase reporter gene. Meth Enzymol 1992, 216: 397-414.
- Millar AJ, Short SR, Hiratsuka K, Chua N-H, Kay SA: Firefly luciferase as a
reporter of regulated gene expression in higher plants. Plant Mol Biol Rep 1992,
10: 324-337.
- Sheen J: Metabolic repression of transcription in higher plants. Plant Cell
1990, 2: 1027-1038.
- Sheen J: Molecular mechanisms underlying the differential expression of
maize pyruvate, orthophosphate dikinase genes. Plant Cell 1991, 3: 225-245.
- Schäffner AR, Sheen J : Maize rbcS promoter activity depends on sequence
elements not found in dicot rbcS promoters. Plant Cell 1991, 3:997-1012.
- Schäffner AR, Sheen J: Maize C4 photosynthesis involves differential
regulation of maize PEPC genes. Plant J 1992, 2: 221-232.
- Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ: Soybean GH3 promoter
contains multiple auxin-inducible elements. Plant Cell 1994, 6: 645-657.
- Röbrig H, Schmidt J, Walden R, Czaja l, Miklasevics E, Wieneke U, Schell J,
John M: Growth of tobacco protoplasts stimulated by synthetic lipo
chitooligosaccharides. Science 1995, 269: 841-843.
- van der Krol AR, Chua N-H: The basic domain of plant B-ZIP proteins facilitates
import of a reporter protein into plant nuclei. Plant Cell 1991, 3: 667-675.
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP: Four genes in
two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small
subunit polypeptides of Arabidopsis. Plant Mol Biol 1988, 11: 745-759.
- Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: ß-glucuronidase as a
sensitive and versatile gene fusion marker in higher plants. EMBO J 1987, 6: 3901
3907.
- Varagona MJ, Schmidt RJ, Raikhel NV: Nuclear localization signal(s) required
for nuclear targeting of the maize regulatory protein opaque-2. Plant Cell 1992, 4:
1213-1227.
- Yamaguchi-Shinozaki K, Shinozaki K: A novel cis-acting element in an
Arabidopsis gene is involved in responsiveness to drought, low-temperature, or
high-salt stress. Plant Cell 1994, 6: 251-264.
- Youvan DC: Green fluorescent pets. Science 1995, 268: 264.
- Gleave AP: A versatile binary vector system with a T-DNA organizational
structure conducive to efficient integration of cloned DNA into the plant genome.
Plant Mol Biol 1992, 20: 1203-1207.
- Nagy JT, Maliga P: Callus induction and plant regeneration from mesophyll
protoplasts cultured in vitro. Z Pflan 1976, 78: 453-455.
- Horsch RB, Fraiey RT, Rogers SG, Sanders PR, Lloyd A, Hoffmann N:
Inheritance of functional foreign genes in plants. Science 1984, 223: 1229-1231.





